Technical Note 13 – The Limitations of DNA Quantification Using DNA Binding Fluorescent Dyes
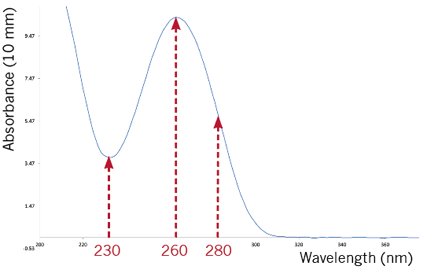
Figure 1: Typical absorbance spectrum scan of a nucleic acid sample
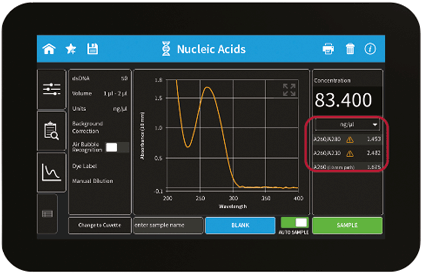
Figure 2: User Interface of the NanoPhotometer® method result screen including sample quality control features (flagged sample for impurities)
Commercially available solutions such as Qubit™ contain fluorescence dyes specifically binding to dsDNA to be measured with a handheld reader. There are different types of kits available depending on the application and the desired concentration range. Fluorescence at first glance seems to be a viable path to accurate dsDNA concentrations for all kinds of samples, but there are limitations that come with this method including intense sample preparation to carry out this relatively costly and time consuming method. dsDNA quantification may be underestimated depending on the method used for the DNA extraction and in case of changing salt concentrations caused by potential dilution steps.
The following limitations should be noted when considering using a fluorescence dye based method for dsDNA concentration determinations:
- Samples measured with fluorescence need to be prepared in the appropriate buffer (preferably Tris or at least 1 mM NaCl), as insufficiently buffered solutions may cause an extension of dsDNA due to increased electrostatic repulsion of negatively charged DNA phosphates which lowers binding properties of fluorescent dyes to the sample.
- Buffers with less than 1 mM NaCl transiently destabilize dsDNA, leading to the second conformational state of dsDNA with a low affinity for fluorescent dye. Dilution of DNA in the absence of NaCl also results in irreversible structural changes to DNA. The presence of a minimum of 1 mM NaCl needs to be maintained for all dilution stages.
- Trizol extractions should be avoided if working with fluorescence as they contain guanidine salts, which denature dsDNA to ssDNA resulting in reduced affinity of the dye leading to lower concentration readings.
- Lower pH of ddH2O may cause denaturation of dsDNA to ssDNA, leading to similar negative effects on the binding properties of fluorescent dyes.
- A combination of using Trizol and low salt/pH worsens the effect.
- Heating of samples above 65 °C should be avoided throughout the workflow as it causes denaturation of dsDNA.
- Sonication, especially of unbuffered solutions, leads to fragmentation of DNA and significantly lower fluorescence signals. For the quantification of fragmented DNA, an equally fragmented standard sample should be used to achieve the most reliable result.
- ≥ 0.1% of SDS and ≥ 0.05% CTAB quenches the dye signal and should be avoided.
- Supercoiled plasmid does not bind fluorescent dyes to the same degree as linearized plasmid.
- A major caveat is that the fluorescent dye binding assays require nucleic acid standards that match the properties of samples being measured.
- Fluorescent dyes underestimate the concentration of double-stranded DNA with a size less than 23 kbp.
- Not suitable for the QC of dye-labeled microarray samples.
| DNA | Relative Fluorescence | NanoPhotometer® |
| Frozen | Suitable* | Recommended |
| FFPE | Recommended | Overestimated |
| Trizol-extracted | Underestimated | Recommended |
| Dye-labeled microarray samples | Not suitable | Recommended |
Table 2. The benefit comparison of relative fluorescence and UV/Vis
| Type of Detection | Relative Fluorescence | UV/VIS |
| Detection of Contaminants | No | Yes |
| Concentration Range | Kit dependent 0.01 – 1000 ng/µl | 1 – 16,500 ng/µl |
| Consumables Needed | Yes | No |
| Price Per Measurement | 0.71 – 1.18 USD | 0 USD |
| Incubation Time | 2 – 5 min per sample | N/A |
| Available Applications | Limited | Numerous |
References
Nakayama Y, Yamaguchi H, Einaga N, Esumi M (2016) Pitfalls of DNA Quantification Using DNA-Binding Fluorescent Dyes and Suggested Solutions. PLoS ONE 11(3): e0150528. doi:10.1371/journal.pone.0150528Qubit™ is a registered trademark of Thermo Fisher Scientific. Implen® is not affiliated, sponsored or endorsed by Thermo Fisher Scientific or Qubit™.