Implen Workflow Insights
Workflow Insights Series
The Hidden Risks in NGS Workflows
In the context of next-generation sequencing (NGS), attention is often directed toward the advanced stages of the workflow, including high-throughput sequencing, variant calling, and complex bioinformatics analysis. But in reality, the success of an entire sequencing run depends on the steps that happen before the library prep begins.
Inaccurate quantification or the presence of contaminants in input material can negatively affect the integrity and reliability of an entire workflow. It can adversely affect library construction efficiency, compromise read coverage, and consequently reduce the number of genes detected.
This is particularly critical in applications involving high-value or limited biological samples, where even minor errors can lead to compromised data, waste of resources, and the need for costly repetition.
Sample Quantification and Purity Matter
Small early errors can affect your NGS results. Here’s what can go wrong:
- Inaccurate concentration measurements can lead to incorrect input amounts, resulting in poor coverage, failed libraries, or biased representation of transcripts (McNulty et al., 2020).
- Contaminants (like proteins, phenol, or residual ethanol) can inhibit enzymes used during reverse transcription or adapter ligation.
- Sample degradation can result in biased transcript detection or incomplete genome coverage (Gallego Romero et al., 2014).
The earlier you catch these problems, the easier—and cheaper—it is to fix them.
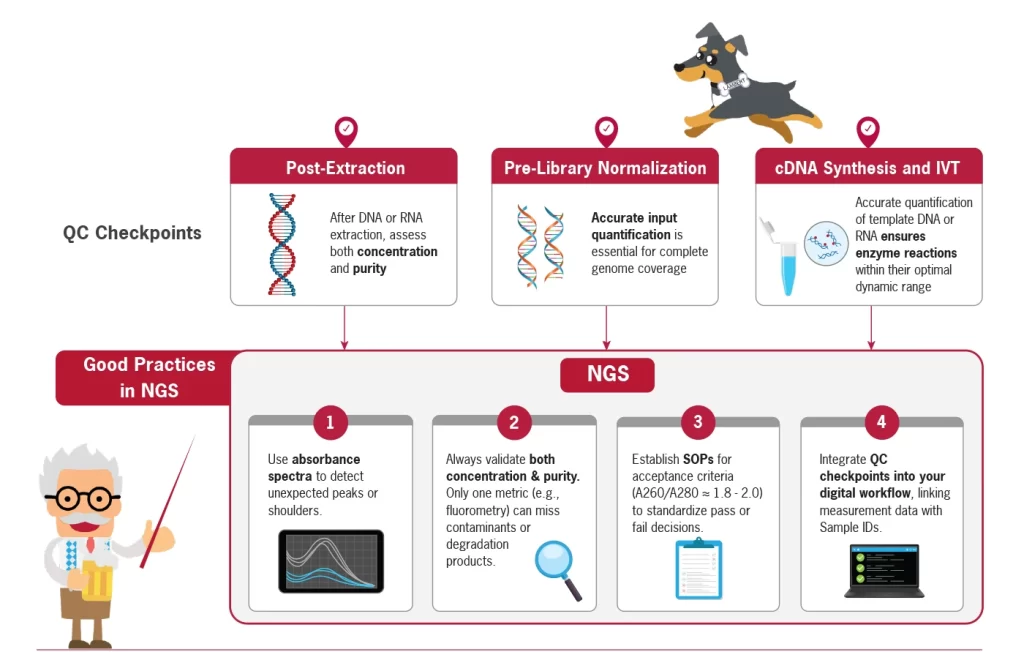
When to Integrate QC
into Your Workflow
- Post-extraction: Immediately after DNA or RNA extraction, assess both concentration and purity. This helps identify degradation, residual solvents (e.g., phenol, ethanol), or protein contamination before investing more time and reagents.
- Pre-library normalization: Before proceeding to library preparation, accurate input quantification is essential for complete genome coverage.
- During cDNA synthesis and IVT (in vitro transcription): Enzymes work best within precise ranges. Therefore, accurate quantification of template DNA or RNA ensures that your reactions are efficient and consistent (Figure 1).
Practical Recommendations
for Your Workflow:
- Use absorbance spectra to detect unexpected peaks or shoulders, revealing hidden impurities (e.g., residual buffers, phenol, or other co-eluted materials).
- Always validate both concentration and purity. Relying solely on one metric (e.g., fluorometry) can miss contaminants or degradation.
- Establish clear SOPs for acceptance criteria (e.g.,
A260/A280 ≈ 1.8–2.0 for DNA, 1.9–2.1 for RNA) to standardize pass or fail decisions. - Integrate QC checkpoints into your digital workflow, linking measurement data with sample IDs to enable traceability (Figure 1).

Conclusion
Whether you are preparing libraries for clinical diagnostics, single-cell transcriptomics, or mRNA-based therapies, remember this: Quality doesn’t start at the sequencer – it starts at the bench.
Integrating solid QC at the beginning of your NGS workflow – supported by reliable analytical tools – ensures:
- High-quality data
- Reliable, reproducible results
- Fewer costly reruns and workflow interruptions
Success of NGS workflows
While fluorometry helps with concentration determination, these assays cannot uncover what’s lurking beneath the surface (like potential contaminating proteins or solvents) or sample degradation.
This is where UV-Vis spectrophotometry steps in. By combining these technologies into your upstream quality control (e.g., Implen NanoPhotometer® and Qubit Fluorometer), you can strengthen the reliability of your downstream outcomes and ensure the overall success of NGS workflows.